30 Inference in R
30.1 BSDA Package
> install.packages("BSDA")> library(BSDA)
> str(z.test)
function (x, y = NULL, alternative = "two.sided", mu = 0, sigma.x = NULL,
sigma.y = NULL, conf.level = 0.95) 30.2 Example: Poisson
Apply z.test():
> set.seed(210)> n <- 40
> lam <- 14
> x <- rpois(n=n, lambda=lam)
> lam.hat <- mean(x)
> stddev <- sqrt(lam.hat)
> z.test(x=x, sigma.x=stddev, mu=lam)
One-sample z-Test
data: x
z = 0.41885, p-value = 0.6753
alternative hypothesis: true mean is not equal to 14
95 percent confidence interval:
13.08016 15.41984
sample estimates:
mean of x
14.25 30.3 Direct Calculations
Confidence interval:
> lam.hat <- mean(x)
> lam.hat
[1] 14.25
> stderr <- sqrt(lam.hat)/sqrt(n)
> lam.hat - abs(qnorm(0.025)) * stderr # lower bound
[1] 13.08016
> lam.hat + abs(qnorm(0.025)) * stderr # upper bound
[1] 15.41984Hypothesis test:
> z <- (lam.hat - lam)/stderr
> z # test statistic
[1] 0.4188539
> 2 * pnorm(-abs(z)) # two-sided p-value
[1] 0.675322930.4 Commonly Used Functions
R has the following functions for doing inference on some of the distributions we have considered.
- Normal:
t.test() - Binomial:
binomial.test()orprop.test() - Poisson:
poisson.test()
These perform one-sample and two-sample hypothesis testing and confidence interval construction for both the one-sided and two-sided cases.
30.5 About These Functions
We covered a convenient, unified MLE framework that allows us to better understand how confidence intervals and hypothesis testing are performed
However, this framework requires large sample sizes and is not necessarily the best method to apply in all circumstances
The above R functions are versatile functions for analyzing Normal, Binomial, and Poisson distributed data (or approximations thereof) that use much broader theory and methods than we have covered
However, the arguments these functions take and the ouput of the functions are in line with the framework that we have covered
30.6 Normal Data: “Davis” Data Set
> library("car")
> data("Davis")> htwt <- tbl_df(Davis)
> htwt
# A tibble: 200 x 5
sex weight height repwt repht
<fct> <int> <int> <int> <int>
1 M 77 182 77 180
2 F 58 161 51 159
3 F 53 161 54 158
4 M 68 177 70 175
5 F 59 157 59 155
6 M 76 170 76 165
7 M 76 167 77 165
8 M 69 186 73 180
9 M 71 178 71 175
10 M 65 171 64 170
# … with 190 more rows30.7 Height vs Weight
> ggplot(htwt) +
+ geom_point(aes(x=height, y=weight, color=sex), size=2, alpha=0.5) +
+ scale_colour_manual(values=c("red", "blue"))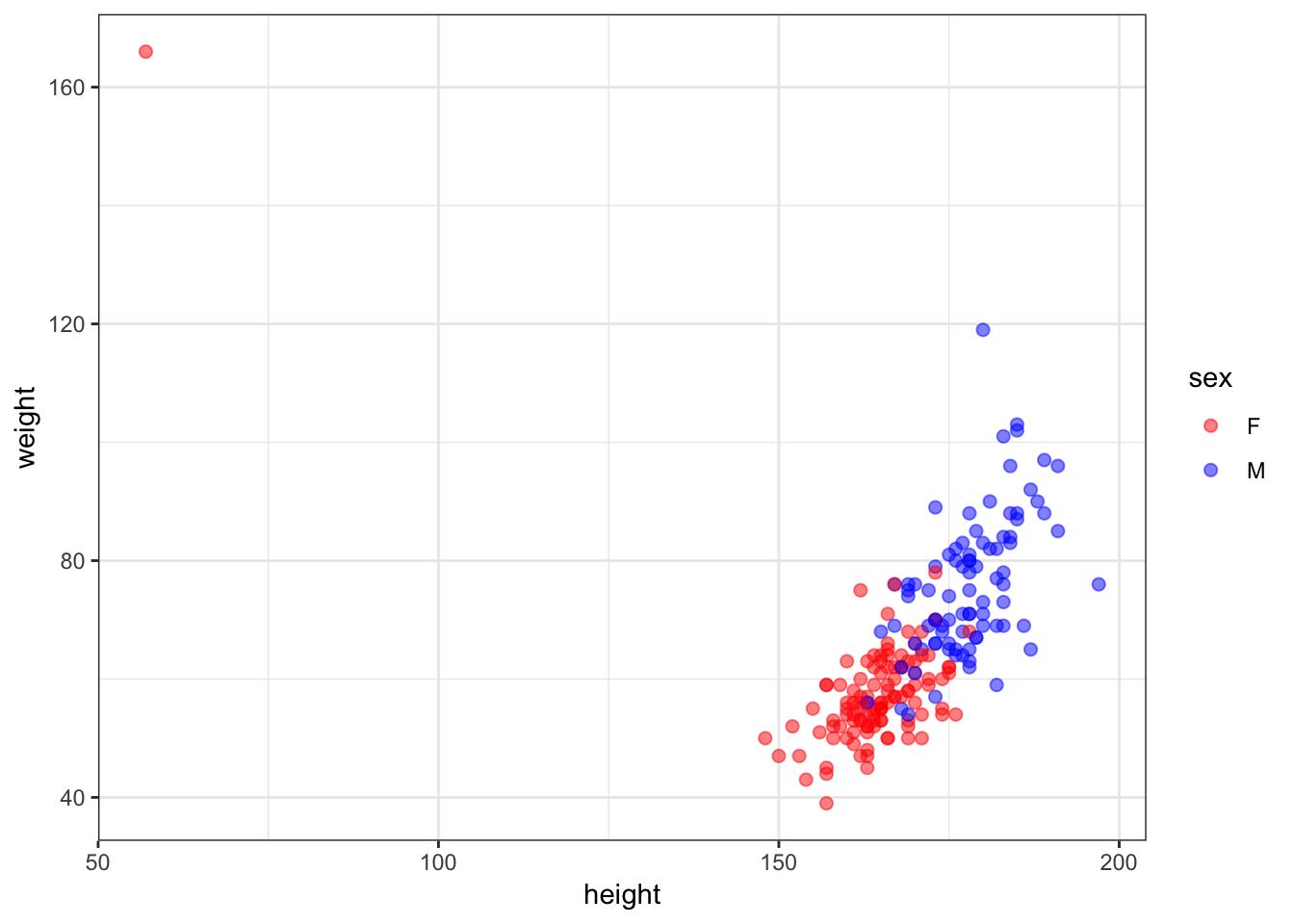
30.8 An Error?
> which(htwt$height < 100)
[1] 12
> htwt[12,]
# A tibble: 1 x 5
sex weight height repwt repht
<fct> <int> <int> <int> <int>
1 F 166 57 56 163> htwt[12,c(2,3)] <- htwt[12,c(3,2)]30.9 Updated Height vs Weight
> ggplot(htwt) +
+ geom_point(aes(x=height, y=weight, color=sex), size=2, alpha=0.5) +
+ scale_color_manual(values=c("red", "blue"))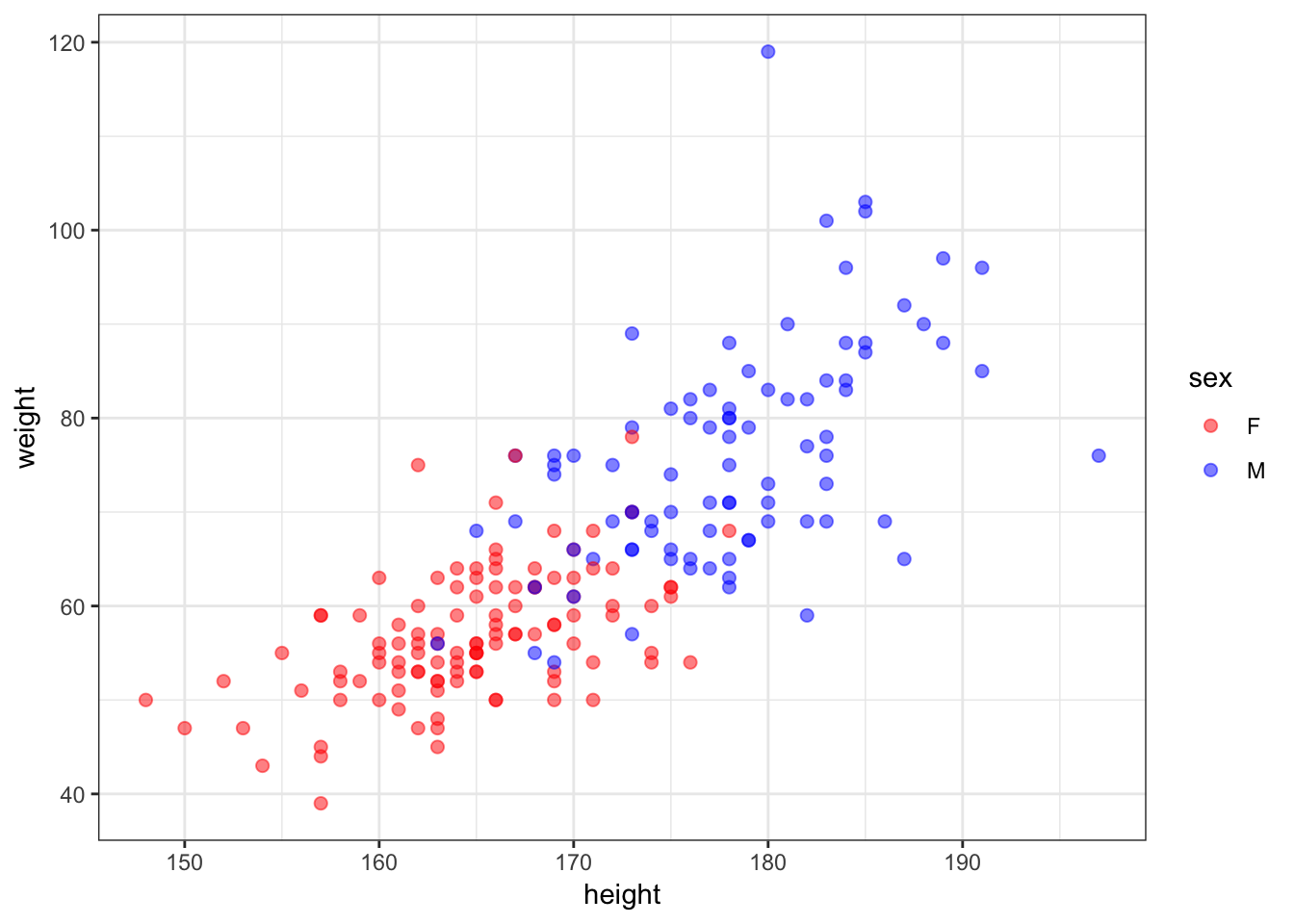
30.10 Density Plots of Height
> ggplot(htwt) +
+ geom_density(aes(x=height, color=sex), size=1.5) +
+ scale_color_manual(values=c("red", "blue"))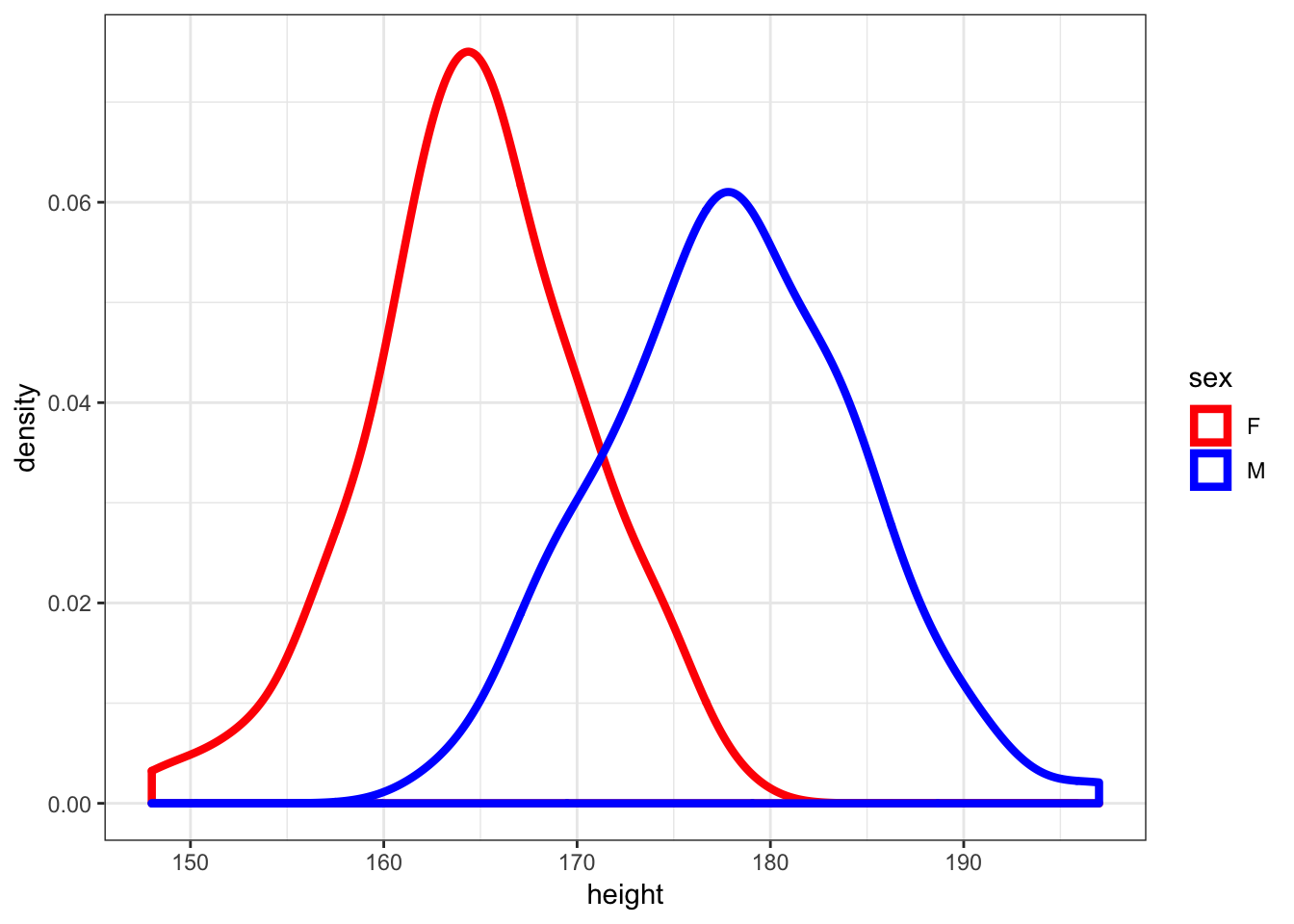
30.11 Density Plots of Weight
> ggplot(htwt) +
+ geom_density(aes(x=weight, color=sex), size=1.5) +
+ scale_color_manual(values=c("red", "blue"))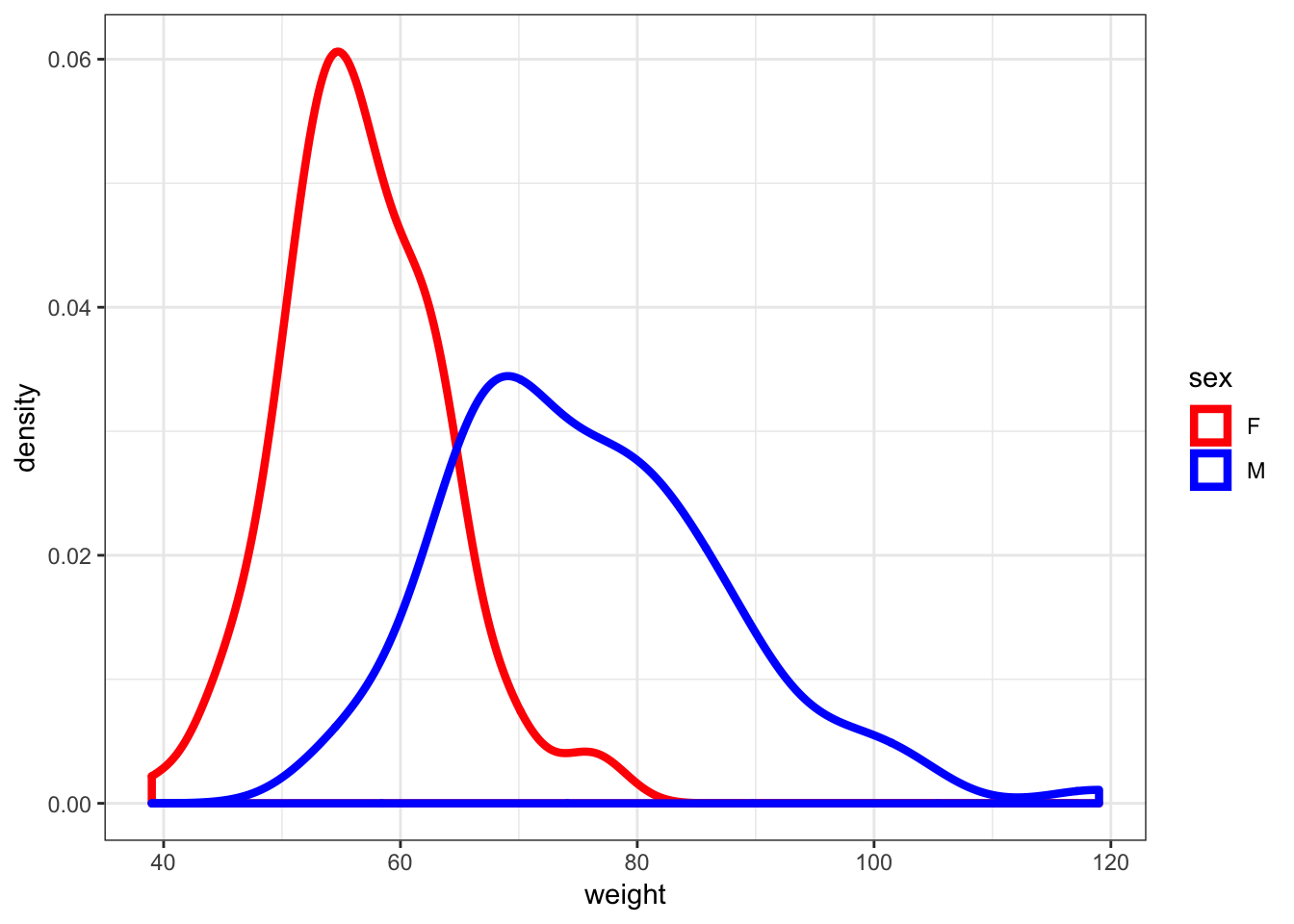
30.12 t.test() Function
From the help file…
Usage
t.test(x, ...)
## Default S3 method:
t.test(x, y = NULL,
alternative = c("two.sided", "less", "greater"),
mu = 0, paired = FALSE, var.equal = FALSE,
conf.level = 0.95, ...)
## S3 method for class 'formula'
t.test(formula, data, subset, na.action, ...)30.13 Two-Sided Test of Male Height
> m_ht <- htwt %>% filter(sex=="M") %>% select(height)
> testresult <- t.test(x = m_ht$height, mu=177)> class(testresult)
[1] "htest"
> is.list(testresult)
[1] TRUE30.14 Output of t.test()
> names(testresult)
[1] "statistic" "parameter" "p.value" "conf.int" "estimate"
[6] "null.value" "stderr" "alternative" "method" "data.name"
> testresult
One Sample t-test
data: m_ht$height
t = 1.473, df = 87, p-value = 0.1443
alternative hypothesis: true mean is not equal to 177
95 percent confidence interval:
176.6467 179.3760
sample estimates:
mean of x
178.0114 30.15 Tidying the Output
> library(broom)
> tidy(testresult)
# A tibble: 1 x 8
estimate statistic p.value parameter conf.low conf.high method
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 178. 1.47 0.144 87 177. 179. One S…
# … with 1 more variable: alternative <chr>30.16 Two-Sided Test of Female Height
> f_ht <- htwt %>% filter(sex=="F") %>% select(height)
> t.test(x = f_ht$height, mu = 164)
One Sample t-test
data: f_ht$height
t = 1.3358, df = 111, p-value = 0.1844
alternative hypothesis: true mean is not equal to 164
95 percent confidence interval:
163.6547 165.7739
sample estimates:
mean of x
164.7143 30.17 Difference of Two Means
> t.test(x = m_ht$height, y = f_ht$height)
Welch Two Sample t-test
data: m_ht$height and f_ht$height
t = 15.28, df = 174.29, p-value < 2.2e-16
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
11.57949 15.01467
sample estimates:
mean of x mean of y
178.0114 164.7143 30.18 Test with Equal Variances
> htwt %>% group_by(sex) %>% summarize(sd(height))
# A tibble: 2 x 2
sex `sd(height)`
<fct> <dbl>
1 F 5.66
2 M 6.44
> t.test(x = m_ht$height, y = f_ht$height, var.equal = TRUE)
Two Sample t-test
data: m_ht$height and f_ht$height
t = 15.519, df = 198, p-value < 2.2e-16
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
11.60735 14.98680
sample estimates:
mean of x mean of y
178.0114 164.7143 30.19 Paired Sample Test (v. 1)
First take the difference between the paired observations. Then apply the one-sample t-test.
> htwt <- htwt %>% mutate(diffwt = (weight - repwt),
+ diffht = (height - repht))
> t.test(x = htwt$diffwt) %>% tidy()
# A tibble: 1 x 8
estimate statistic p.value parameter conf.low conf.high method
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 0.00546 0.0319 0.975 182 -0.332 0.343 One S…
# … with 1 more variable: alternative <chr>
> t.test(x = htwt$diffht) %>% tidy()
# A tibble: 1 x 8
estimate statistic p.value parameter conf.low conf.high method
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 2.08 13.5 2.64e-29 182 1.77 2.38 One S…
# … with 1 more variable: alternative <chr>30.20 Paired Sample Test (v. 2)
Enter each sample into the t.test() function, but use the paired=TRUE argument. This is operationally equivalent to the previous version.
> t.test(x=htwt$weight, y=htwt$repwt, paired=TRUE) %>% tidy()
# A tibble: 1 x 8
estimate statistic p.value parameter conf.low conf.high method
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 0.00546 0.0319 0.975 182 -0.332 0.343 Paire…
# … with 1 more variable: alternative <chr>
> t.test(x=htwt$height, y=htwt$repht, paired=TRUE) %>% tidy()
# A tibble: 1 x 8
estimate statistic p.value parameter conf.low conf.high method
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 2.08 13.5 2.64e-29 182 1.77 2.38 Paire…
# … with 1 more variable: alternative <chr>
> htwt %>% select(height, repht) %>% na.omit() %>%
+ summarize(mean(height), mean(repht))
# A tibble: 1 x 2
`mean(height)` `mean(repht)`
<dbl> <dbl>
1 171. 168.30.21 The Coin Flip Example
I flip it 20 times and it lands on heads 16 times.
- My data is \(x=16\) heads out of \(n=20\) flips.
- My data generation model is \(X \sim \mbox{Binomial}(20, p)\).
- I form the statistic \(\hat{p} = 16/20\) as an estimate of \(p\).
Let’s do hypothesis testing and confidence interval construction on these data.
30.22 binom.test()
> str(binom.test)
function (x, n, p = 0.5, alternative = c("two.sided", "less", "greater"),
conf.level = 0.95)
> binom.test(x=16, n=20, p = 0.5)
Exact binomial test
data: 16 and 20
number of successes = 16, number of trials = 20, p-value = 0.01182
alternative hypothesis: true probability of success is not equal to 0.5
95 percent confidence interval:
0.563386 0.942666
sample estimates:
probability of success
0.8 30.23 alternative = "greater"
Tests \(H_0: p \leq 0.5\) vs. \(H_1: p > 0.5\).
> binom.test(x=16, n=20, p = 0.5, alternative="greater")
Exact binomial test
data: 16 and 20
number of successes = 16, number of trials = 20, p-value =
0.005909
alternative hypothesis: true probability of success is greater than 0.5
95 percent confidence interval:
0.5989719 1.0000000
sample estimates:
probability of success
0.8 30.24 alternative = "less"
Tests \(H_0: p \geq 0.5\) vs. \(H_1: p < 0.5\).
> binom.test(x=16, n=20, p = 0.5, alternative="less")
Exact binomial test
data: 16 and 20
number of successes = 16, number of trials = 20, p-value = 0.9987
alternative hypothesis: true probability of success is less than 0.5
95 percent confidence interval:
0.0000000 0.9286461
sample estimates:
probability of success
0.8 30.25 prop.test()
This is a “large \(n\)” inference method that is very similar to our \(z\)-statistic approach.
> str(prop.test)
function (x, n, p = NULL, alternative = c("two.sided", "less", "greater"),
conf.level = 0.95, correct = TRUE)
> prop.test(x=16, n=20, p=0.5)
1-sample proportions test with continuity correction
data: 16 out of 20, null probability 0.5
X-squared = 6.05, df = 1, p-value = 0.01391
alternative hypothesis: true p is not equal to 0.5
95 percent confidence interval:
0.5573138 0.9338938
sample estimates:
p
0.8 30.26 An Observation
> p <- binom.test(x=16, n=20, p = 0.5)$p.value
> binom.test(x=16, n=20, p = 0.5, conf.level=(1-p))
Exact binomial test
data: 16 and 20
number of successes = 16, number of trials = 20, p-value = 0.01182
alternative hypothesis: true probability of success is not equal to 0.5
98.81821 percent confidence interval:
0.5000000 0.9625097
sample estimates:
probability of success
0.8 Exercise: Figure out what happened here.
30.27 Wording of Surveys
The way a question is phrased can influence a person’s response. For example, Pew Research Center conducted a survey with the following question:
“As you may know, by 2014 nearly all Americans will be required to have health insurance. [People who do not buy insurance will pay a penalty] while [People who cannot afford it will receive financial help from the government]. Do you approve or disapprove of this policy?”
For each randomly sampled respondent, the statements in brackets were randomized: either they were kept in the order given above, or the two statements were reversed.
Credit: This example comes from Open Intro Statistics, Exercise 6.10.
30.28 The Data
The following table shows the results of this experiment.
| 2nd Statement | Sample Size | Approve Law | Disapprove Law | Other |
|---|---|---|---|---|
| “people who cannot afford it will receive financial help from the government” | 771 | 47 | 49 | 3 |
| “people who do not buy it will pay a penalty” | 732 | 34 | 63 | 3 |
30.29 Inference on the Difference
Create and interpret a 90% confidence interval of the difference in approval. Also perform a hyppthesis test that the approval rates are equal.
> x <- round(c(0.47*771, 0.34*732))
> n <- round(c(771*0.97, 732*0.97))
> prop.test(x=x, n=n, conf.level=0.90)
2-sample test for equality of proportions with continuity
correction
data: x out of n
X-squared = 26.023, df = 1, p-value = 3.374e-07
alternative hypothesis: two.sided
90 percent confidence interval:
0.08979649 0.17670950
sample estimates:
prop 1 prop 2
0.4839572 0.3507042 30.30 90% Confidence Interval
Let’s use MLE theory to construct of a two-sided 90% CI.
> p1.hat <- 0.47
> n1 <- 771
> p2.hat <- 0.34
> n2 <- 732
> stderr <- sqrt(p1.hat*(1-p1.hat)/n1 + p2.hat*(1-p2.hat)/n2)
>
> # the 90% CI
> (p1.hat - p2.hat) + c(-1,1)*abs(qnorm(0.05))*stderr
[1] 0.08872616 0.1712738430.31 Poisson Data: poisson.test()
> str(poisson.test)
function (x, T = 1, r = 1, alternative = c("two.sided", "less", "greater"),
conf.level = 0.95) From the help:
Arguments
x number of events. A vector of length one or two.
T time base for event count. A vector of length one or two.
r hypothesized rate or rate ratio
alternative indicates the alternative hypothesis and must be one of
"two.sided", "greater" or "less". You can specify just the initial letter.
conf.level confidence level for the returned confidence interval.30.32 Example: RNA-Seq
RNA-Seq gene expression was measured for p53 lung tissue in 12 healthy individuals and 14 individuals with lung cancer.
The counts were given as follows.
Healthy: 82 64 66 88 65 81 85 87 60 79 80 72
Cancer: 59 50 60 60 78 69 70 67 72 66 66 68 54 62
It is hypothesized that p53 expression is higher in healthy individuals. Test this hypothesis, and form a 99% CI.
30.33 \(H_1: \lambda_1 \not= \lambda_2\)
> healthy <- c(82, 64, 66, 88, 65, 81, 85, 87, 60, 79, 80, 72)
> cancer <- c(59, 50, 60, 60, 78, 69, 70, 67, 72, 66, 66, 68,
+ 54, 62)> poisson.test(x=c(sum(healthy), sum(cancer)), T=c(12,14),
+ conf.level=0.99)
Comparison of Poisson rates
data: c(sum(healthy), sum(cancer)) time base: c(12, 14)
count1 = 909, expected count1 = 835.38, p-value = 0.0005739
alternative hypothesis: true rate ratio is not equal to 1
99 percent confidence interval:
1.041626 1.330051
sample estimates:
rate ratio
1.177026 30.34 \(H_1: \lambda_1 < \lambda_2\)
> poisson.test(x=c(sum(healthy), sum(cancer)), T=c(12,14),
+ alternative="less", conf.level=0.99)
Comparison of Poisson rates
data: c(sum(healthy), sum(cancer)) time base: c(12, 14)
count1 = 909, expected count1 = 835.38, p-value = 0.9998
alternative hypothesis: true rate ratio is less than 1
99 percent confidence interval:
0.000000 1.314529
sample estimates:
rate ratio
1.177026 30.35 \(H_1: \lambda_1 > \lambda_2\)
> poisson.test(x=c(sum(healthy), sum(cancer)), T=c(12,14),
+ alternative="greater", conf.level=0.99)
Comparison of Poisson rates
data: c(sum(healthy), sum(cancer)) time base: c(12, 14)
count1 = 909, expected count1 = 835.38, p-value = 0.0002881
alternative hypothesis: true rate ratio is greater than 1
99 percent confidence interval:
1.053921 Inf
sample estimates:
rate ratio
1.177026 30.36 Question
Which analysis is the more informative and scientifically correct one, and why?